
|
||||
| Home | Services | CDISC | Software | About us |
With a new standard for the exchange of clinical data now available (the CDISC standard, not surprisingly being written in XML), CROs and pharma sponsor companies can now save enormous amounts of money (and even more important: time-to-market) in costs of data exchange. The establishment of the new standard also enables CROs and sponsors to do their information exchange much more efficiently.
Imagine a sponsor wants to receive updated clinical data from a CRO on a weekly basis. The CRO has to do so for many of its projects with different pharma companies. As it works now, the CRO will weekly collect all necessary data and 'burn' a CD and send them by parcel service to the sponsor. Or the data may be send by e-mail (security !?).
If the CRO makes a web service available to all its sponsors, then each of these sponsors can make a small application that weekly connects to the web service, which collects the data from the database, transforms them into the CDISC format, and sends the information back as a SOAP response.
As SOAP is able to use HTTPS (secure HTTP), authentication, encryption and electronic signatures (presumable XMLSignature will be used), such a web service can be made available in a very secure way. Essentially, the information exchange can be fully automated. |  | |
 | ||
 |
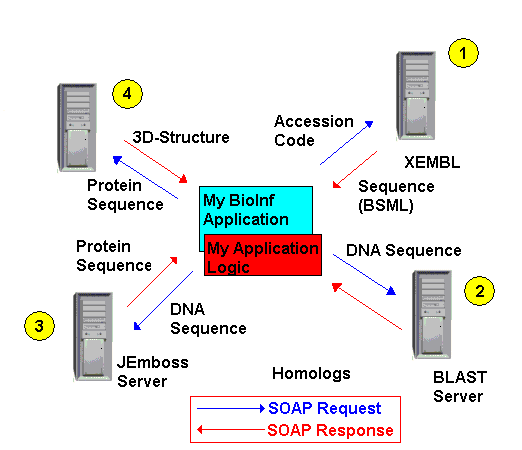
If there is something a bioinformatician does not have, it's time! With the amounts of data becoming available daily, a bioinformatician does not want to upload data to a webserver using a browser, to do certain calculations on a remote machine. Instead he or she wants to automate this, and writes a script for this. This is very easy if the remote server makes it application methods ("services") available as a SOAP service.
Imagine the following process: Starting from a series of DNA accession codes, a bioinformatician
wants to find all available homologs and predict the 3D structure of the resulting protein. His (or here) application
subsequently calls the methods of the XEMBL server
at the EBI (an example Perl script -as provided by EBI- can be
found here - an example Java file is also available on the XEMBL website), sends the
accession code(s) and gets DNA sequences back. These sequences can now be treated within the users own application.
In the next step, the DNA sequences have to be compared with a database of available sequences, using a BLAST alignment.
To do so, the users application sends each sequence to a BLAST web service (there are several of them), which returns
a set of homologs. These can again be treated by the users own application logic.
If now, the user wants each of these DNA sequences be translated into an amino acid sequence, the DNA sequence(s)will
be send to a web service like the JEmboss server.
The result of the JEmboss server is a protein amino acid sequence.
If one wants to develop new drugs that interact with a protein however, one needs to know the proteins 3-dimensional structure.
Doing this experimentally, can be a matter of several years of work, so if a protein is known with a sequence that
is not too different, one can use "homology modelling" to obtain a reasonable 3D-model for that protein. Typically,
one submits the amino acid sequence, using a web browser, to SwissModel.
In future, SwissModel will presumable also become available as a web service, enabling the user to submit the sequence from
within his/her own application.
Insteading of using "copy-and-paste" and/or uploading files using a web browser, Web Services enable to fully automate the process. Once the application (which can be a simple Perl script) written and tested, the user can start the application, and go home or go drinking a coffee (most bioinformaticians however will start doing some other useful work).
More and more, the FDA wants to receive data in a standardized way.
In practice, this means that the data are
provided in XML format (e.g. CDISC data) or in XML format (the backbone - containing the meta information) with
the real information as PDF documents declared as 'leaf elements' in the XML backbone (eCTD).
The FDA has already developed AERS (Adverse Event Reporting System), where consumers, health professionals and others
can submit information about observed adverse events of drugs through the use of a website. The information is then
stored in a database, and evaluated by FDA specialists.
For submitting larger amounts of information, such as CDISC ODM files, or for electronic submissions of NDA's, however,
this mechanism is no longer feasible. Therefore, experiments with web services are foreseen. These will probably
be based on ebXML, which is build on top of SOAP (one can
also say that it is a SOAP extension).
This will make the communication between the FDA and pharma companies much more automated and interactively. We
will not be surprised if in future, the FDA will not only require 'presentations' (PDF) of 'elaborated' data, but the
original raw data (in XML format) itself. Web services are then the ideal way of an automated way of communicating these data.
